21.1 Viral Evolution, Morphology, and Classification
Learning Outcomes
- Describe the general structure of a virus
- Recognize the basic shapes of viruses
- Describe the basis for the Baltimore classification system
- Use the Baltimore classification system to organize viruses into classes
Discovery and Detection
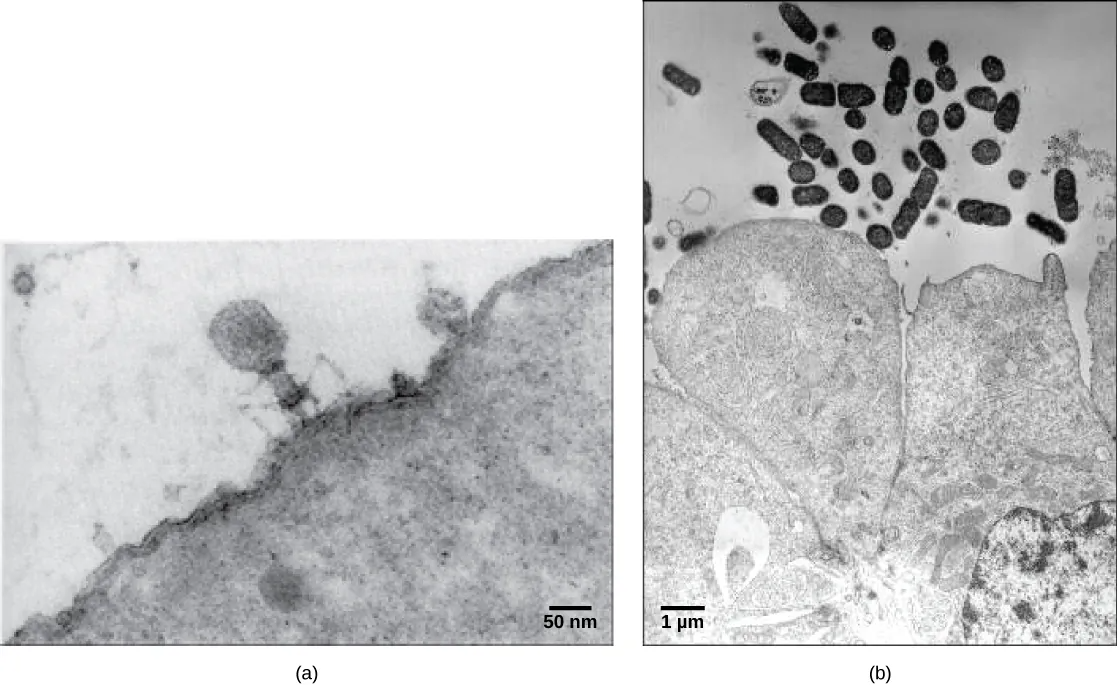
Viruses were first discovered after the development of a porcelain filter—the Chamberland-Pasteur filter—that could remove all bacteria visible in the microscope from any liquid sample. In 1886, Adolph Meyer demonstrated that a disease of tobacco plants—tobacco mosaic disease—could be transferred from a diseased plant to a healthy one via liquid plant extracts. In 1892, Dmitri Ivanowski showed that this disease could be transmitted in this way even after the Chamberland-Pasteur filter had removed all viable bacteria from the extract. Still, it was many years before it was proved that these “filterable” infectious agents were not simply very small bacteria but were a new type of very small, disease-causing particle.
Most virions, or single virus particles, are very small, about 20 to 250 nanometers in diameter. However, some recently discovered viruses from amoebae range up to 1000 nm in diameter. With the exception of large virions, like the poxvirus and other large DNA viruses, viruses cannot be seen with a light microscope. It was not until the development of the electron microscope in the late 1930s that scientists got their first good view of the structure of the tobacco mosaic virus (TMV) (Figure 21.1), and other viruses (Figure 21.2).
Viral Morphology
Viruses are noncellular, meaning they are biological entities that do not have a cellular structure. They, therefore, lack most of the components of cells, such as organelles, ribosomes, and the plasma membrane. A virion consists of a nucleic acid core, an outer protein coating or capsid, and sometimes an outer envelope made of protein and phospholipid membranes derived from the host cell. Viruses may also contain additional proteins, such as enzymes, within the capsid or attached to the viral genome. The most obvious difference between members of different viral families is the variation in their morphology, which is quite diverse.
Morphology
Viruses come in many shapes and sizes, but these features are consistent for each viral family. As we have seen, all virions have a nucleic acid genome covered by a protective capsid. The proteins of the capsid are encoded in the viral genome, and are called capsomeres. Some viral capsids are simple helices or polyhedral “spheres,” whereas others are quite complex in structure (Figure 21.3).
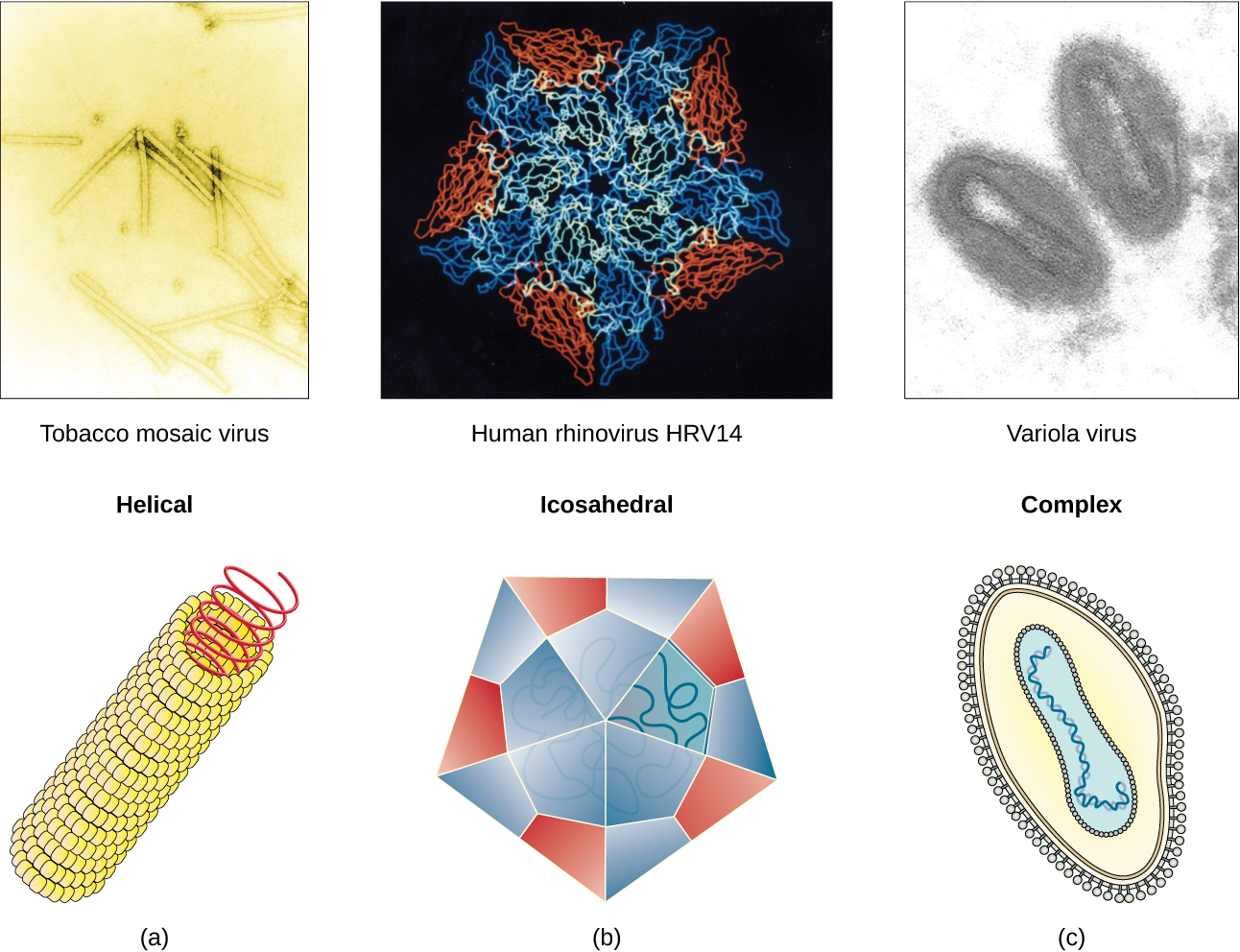
In general, the capsids of viruses are classified into four groups:
- helical – Helical capsids are long and cylindrical. Many plant viruses are helical, including TMV.
- icosahedral – Icosahedral viruses have shapes that are roughly spherical, such as those of poliovirus or herpesviruses.
- enveloped – Enveloped viruses have membranes derived from the host cell that surrounds the capsids. Animal viruses, such as HIV, are frequently enveloped.
- head-and-tail – Head-and-tail viruses infect bacteria and have a head that is similar to icosahedral viruses and a tail shaped like helical viruses.
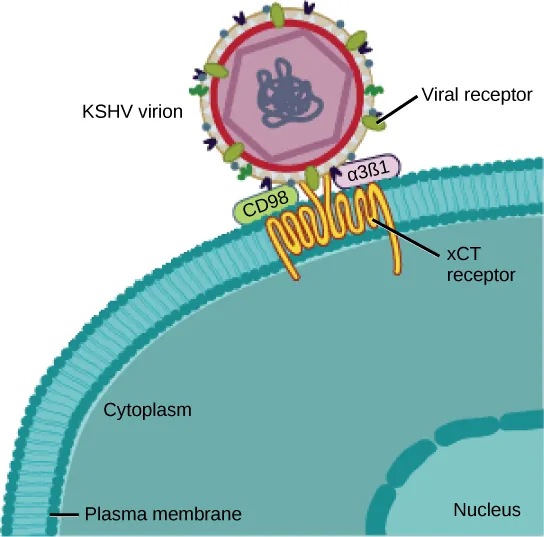
Many viruses use some sort of glycoprotein to attach to their host cells via molecules on the cell called viral receptors. For these viruses, attachment is required for later penetration of the cell membrane; only after penetration takes place can the virus complete its replication inside the cell. The receptors that viruses use are molecules that are normally found on cell surfaces and have their own physiological functions. It appears that viruses have simply evolved to make use of these molecules for their own replication. For example, HIV uses the CD4 molecule on T lymphocytes as one of its receptors (Figure 21.4). CD4 is a type of molecule called a cell adhesion molecule, which functions to keep different types of immune cells in close proximity to each other during the generation of a T lymphocyte immune response.
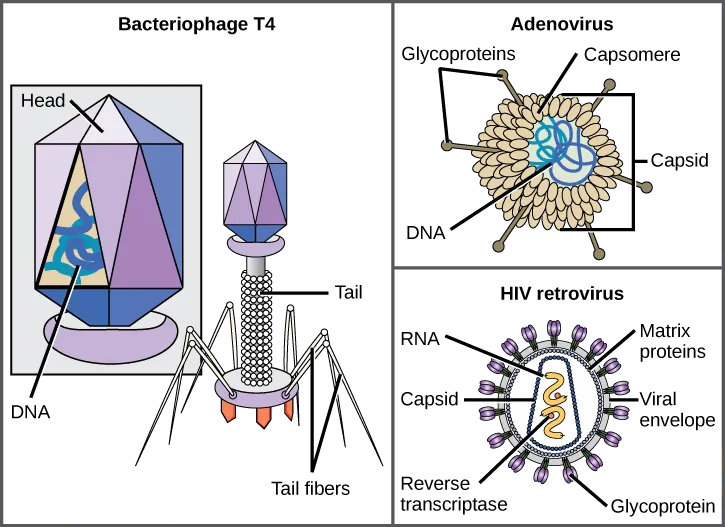
One of the most complex virions known, the T4 bacteriophage (which infects the Escherichia coli) bacterium, has a tail structure that the virus uses to attach to host cells and a head structure that houses its DNA.
Adenovirus, a non-enveloped animal virus that causes respiratory illnesses in humans, uses glycoprotein spikes protruding from its capsomeres to attach to host cells. Non-enveloped viruses also include those that cause polio (poliovirus), plantar warts (papillomavirus), and hepatitis A (hepatitis A virus).
Enveloped virions, such as the influenza virus, consist of nucleic acid (RNA in the case of influenza) and capsid proteins surrounded by a phospholipid bilayer envelope that contains virus-encoded proteins. Glycoproteins embedded in the viral envelope are used to attach to host cells. Other envelope proteins are the matrix proteins that stabilize the envelope and often play a role in the assembly of progeny virions. Chicken pox, HIV, and mumps are other examples of diseases caused by viruses with envelopes. Because of the fragility of the envelope, non-enveloped viruses are more resistant to changes in temperature, pH, and some disinfectants than enveloped viruses.
Note: for this course, there is no need to memorize what type of virus each of these examples are. They are only mentioned to illustrate the diversity of viruses that you might have heard about.
Overall, the shape of the virion and the presence or absence of an envelope tell us little about what disease the virus may cause or what species it might infect, but they are still useful means to begin viral classification (Figure 21.5).
Types of Nucleic Acid
Unlike nearly all living organisms that use DNA as their genetic material, viruses may use either DNA or RNA. The virus core contains the genome—the total genetic content of the virus. Viral genomes tend to be small, containing only those genes that encode proteins that the virus cannot get from the host cell. This genetic material may be single- or double-stranded. It may also be linear or circular. While most viruses contain a single nucleic acid, others have genomes divided into several segments. The RNA genome of the influenza virus is segmented, which contributes to its variability and continuous evolution, and explains why it is difficult to develop a vaccine against it.
In DNA viruses, the viral DNA directs the host cell’s replication proteins to synthesize new copies of the viral genome and to transcribe and translate that genome into viral proteins. Human diseases caused by DNA viruses include chickenpox, hepatitis B, and adenoviruses. Sexually transmitted DNA viruses include the herpes virus and the human papilloma virus (HPV), which has been associated with cervical cancer and genital warts.
RNA viruses contain only RNA as their genetic material. To replicate their genomes in the host cell, the RNA viruses must encode their own enzymes that can replicate RNA into RNA or, in the retroviruses, into DNA. These RNA polymerase enzymes are more likely to make copying errors than DNA polymerases, and therefore often make mistakes during transcription. For this reason, mutations in RNA viruses occur more frequently than in DNA viruses. This causes them to change and adapt more rapidly to their host. Human diseases caused by RNA viruses include influenza, hepatitis C, measles, and rabies. The HIV virus, which is sexually transmitted, is an RNA retrovirus.
Note: for this course, there is no need to memorize what type of virus each of these examples are. They are only mentioned to illustrate the diversity of viruses that you might have heard about.
The Challenge of Virus Classification
Because most viruses probably evolved from different ancestors, the systematic methods that scientists have used to classify prokaryotic and eukaryotic cells are not very useful. Why?, Because viruses have no common genomic sequence that they all share. Biologists have used several classification systems in the past. Viruses were initially grouped by shared morphology. Later, groups of viruses were classified by the type of nucleic acid they contained, DNA or RNA, and whether their nucleic acid was single- or double-stranded. However, these earlier classification methods grouped viruses differently, because they were based on different sets of characters of the virus. The most commonly used classification method today is called the Baltimore classification scheme.
The Baltimore classification scheme groups viruses according to how the mRNA is produced during the replicative cycle of the virus.
Group I viruses contain double-stranded DNA (dsDNA) as their genome. Their mRNA is produced by transcription in much the same way as with cellular DNA, using the enzymes of the host cell.
Group II virusesGroup II viruses have single-stranded DNA (ssDNA) as their genome. They convert their single-stranded genomes into a dsDNA intermediate before transcription to mRNA can occur.
Group III viruses use dsRNA as their genome. The strands separate, and one of them is used as a template for the generation of mRNA using the RNA-dependent RNA polymerase encoded by the virus.
Group IV viruses have ssRNA as their genome with a positive polarity, which means that the genomic RNA can serve directly as mRNA. Intermediates of dsRNA, called replicative intermediates, are made in the process of copying the genomic RNA. Multiple, full-length RNA strands of negative polarity (complementary to the positive-stranded genomic RNA) are formed from these intermediates, which may then serve as templates for the production of RNA with positive polarity, including both full-length genomic RNA and shorter viral mRNAs.
Group V viruses contain ssRNA genomes with a negative polarity, meaning that their sequence is complementary to the mRNA. As with Group IV viruses, dsRNA intermediates are used to make copies of the genome and produce mRNA. In this case, the negative-stranded genome can be converted directly to mRNA. Additionally, full-length positive RNA strands are made to serve as templates for the production of the negative-stranded genome.
Group VI viruses have diploid (two copies) ssRNA genomes that must be converted, using the enzyme reverse transcriptase, to dsDNA; the dsDNA is then transported to the nucleus of the host cell and inserted into the host genome. Then, mRNA can be produced by transcription of the viral DNA that was integrated into the host genome.
Group VII viruses have partial dsDNA genomes and make ssRNA intermediates that act as mRNA, but are also converted back into dsDNA genomes by reverse transcriptase, necessary for genome replication.
The characteristics of each group in the Baltimore classification are summarized in Table 21.3. with examples of each group.
| Baltimore Classification | |||
|---|---|---|---|
| Group | Characteristics | Mode of mRNA Production | Example |
| I | Double-stranded DNA | mRNA is transcribed directly from the DNA template | Herpes simplex (herpesvirus) |
| II | Single-stranded DNA | DNA is converted to double-stranded form before RNA is transcribed | Canine parvovirus (parvovirus) |
| III | Double-stranded RNA | mRNA is transcribed from the RNA genome | Childhood gastroenteritis (rotavirus) |
| IV | Single stranded RNA (+) | Genome functions as mRNA | Common cold (picornavirus) |
| V | Single stranded RNA (-) | mRNA is transcribed from the RNA genome | Rabies (rhabdovirus) |
| VI | Single stranded RNA viruses with reverse transcriptase | Reverse transcriptase makes DNA from the RNA genome; DNA is then incorporated in the host genome; mRNA is transcribed from the incorporated DNA | Human immunodeficiency virus (HIV) |
| VII | Double stranded DNA viruses with reverse transcriptase | The viral genome is double-stranded DNA, but viral DNA is replicated through an RNA intermediate; the RNA may serve directly as mRNA or as a template to make mRNA | Hepatitis B virus
(hepadnavirus) |
combination of carbohydrates and proteins
virus with a dsDNA genome
virus with an ssDNA genome
virus with a dsRNA genome
virus with an ssRNA genome with positive polarity
virus with an ssRNA genome with negative polarity
virus with an ssRNA genome converted into dsDNA by reverse transcriptase
virus with a single-stranded mRNA converted into dsDNA for genome replication

